Phosphoswitch-substrate targeting: the dynamic control of the p90 ribosomal S6 kinase family
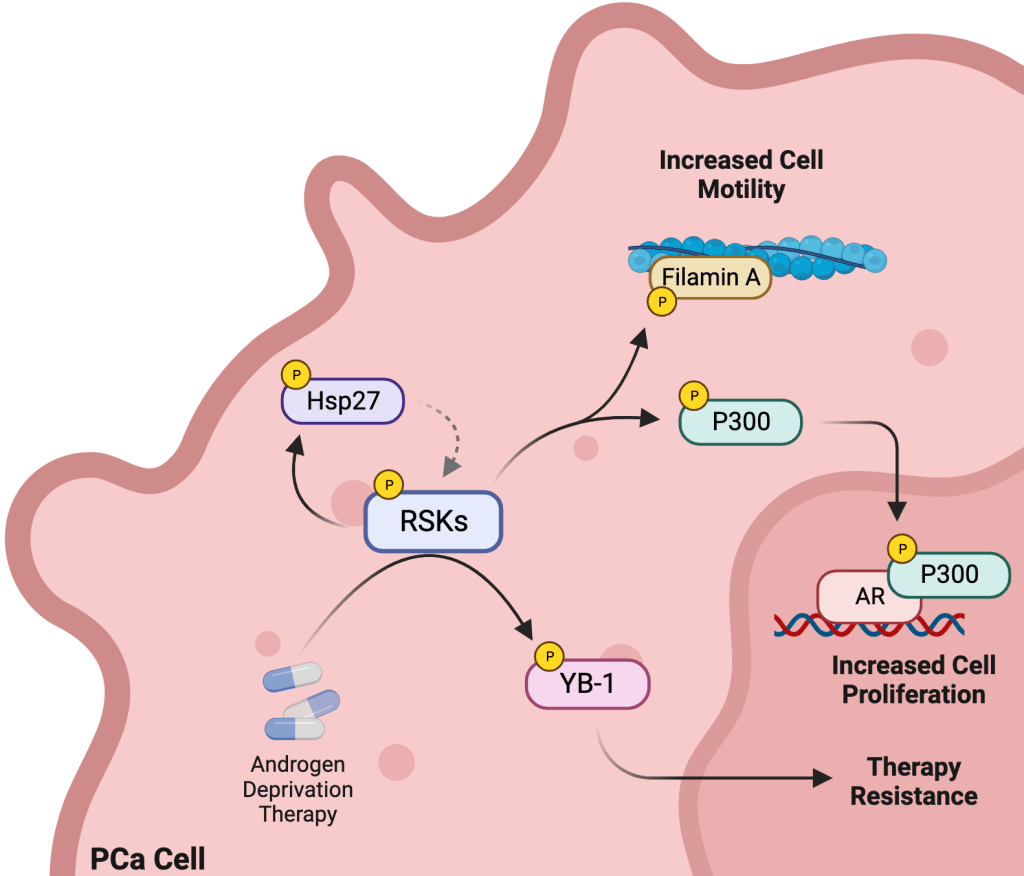
The p90 ribosomal S6 kinase (RSKs) family is a group of ERK/MAPK effectors, which modulates cell transformation, tumorigenesis, and metastasis by phosphorylating both nuclear and cytoplasmic targets. An increasing number of studies have shown that the four isoforms in humans (RSK1-4) have different and specific roles. For example, we have shown that silencing RSK4, but not RSK1, in lung and bladder cancer cell sensitized tumor cells to chemotherapy (Sci Transl Med, 2021). We have recently shown that in Prostate Cancer, RSKs levels are increased, and this correlated with enhanced Androgen Receptor (AR) activity. However, significant gaps remain in our knowledge of how the RSKs are activated and how this impacts down-stream signalling. We aim to understand which unique transcriptional and translational regulators are differentially activated by RSKs, and how this promotes prostate tumorigenesis, invasive progression and drug resistance. We also aim to structurally characterize RSKs complexes and use these data to design protein-protein interaction (PPI) inhibitors that block RSKs downstream signaling by preventing substrate binding.
Collaboration with Dr Greg Brooke (University of Essex) and Dr Oliver Pardo (Imperial College London).

Phosphoregulation of DNA/RNA-binding proteins
Phosphorylation of DNA/RNA-binding proteins by kinases is an important on/off switch to control gene transcription, mRNA splicing and mRNA decay in response to cellular signals. Deregulation of these processes is associated with several human diseases, including cancer and neurodegenerative disorders. Characterising the structural changes induced by phosphorylation is key to provide a mechanistic understanding of DNA/RNA–protein interactions in ribonucleoprotein complexes. We are studying two prototypical DNA/RNA-binding proteins, Y-box binding protein 1 (YB-1 or YBX1) and heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1). We have shown that both proteins can be phosphorylated on several different residues by the ribosomal kinases (S6Ks and RSKs). Our aim is to understand at a structural and functional level the role different phosphorylations play in DNA/RNA recognition and binding specificity.
Collaboration with Dr Vincenzo Venditti (Iowa State University) and Dr Konstantin Roeder (King’s College London).
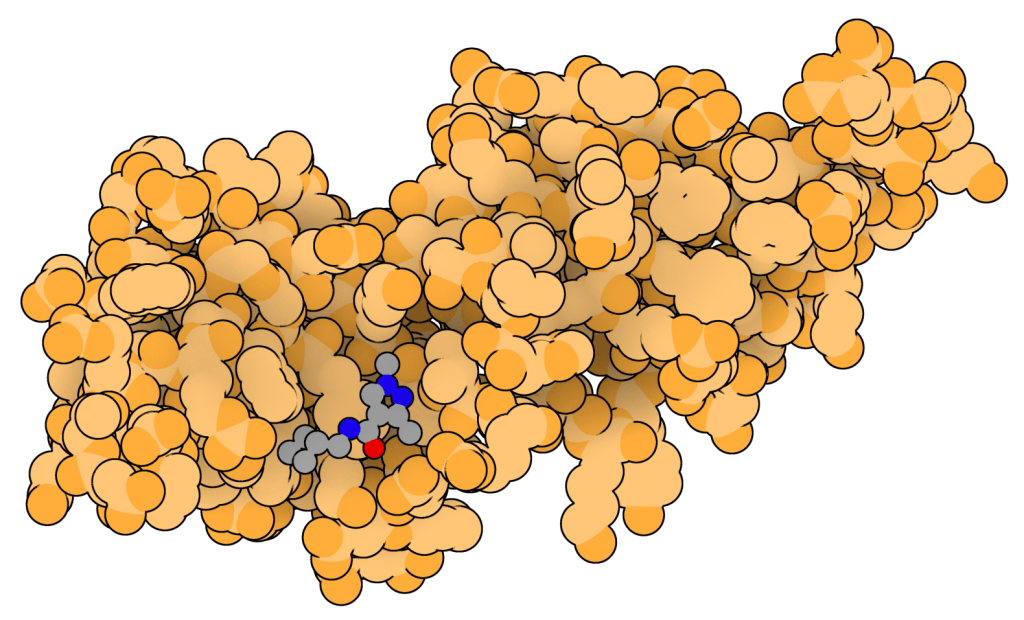
Development of specific inhibitors that prevent hnRNPA1-mediated therapy resistance in Small Cell Lung Cancer
The heterogeneous nuclear ribonucleoprotein (hnRNP) A1 protein holds promises as a novel Small Cell Lung Cancer (SCLC) therapeutic target. hnRNPA1 is a major regulator of gene expression, and misregulation is often associated with cancer. Specifically in SCLC, it has been shown that, downstream of FGF2 signalling, a multi-protein complex involving PKCε, B-Raf and S6K2 is formed in a MEK-dependent fashion. This results in S6K2 activation leading to hnRNPA1 phosphorylation on Ser 4 and Ser 6. This event confers hnRNPA1 the ability to specifically bind the XIAP and Bcl-xL mRNAs. hnRNPA1 mediates nuclear export of these mRNAs and promotes their translation into proteins, leading to downregulation of apoptosis and promotion of cell survival, which unavoidably leads to chemoresistance.
We carried out a fragment-based screening on hnRNPA1 using the X-Chem facility. We are currently developing bioinformatics and AI approaches (J. Chem. Inf. Model., 2024) coupled with our X-Ray structures of hnRNPA1 bound to 35 different fragments (JBC, 2025) to design novel molecules able to prevent hnRNPA1 function in cancer.
Collaboration with Dr Oliver Pardo (Imperial College London), Prof Miraz Rahman (King’s College London) and Prof Tristan Cazenave (Université Paris-Dauphine).
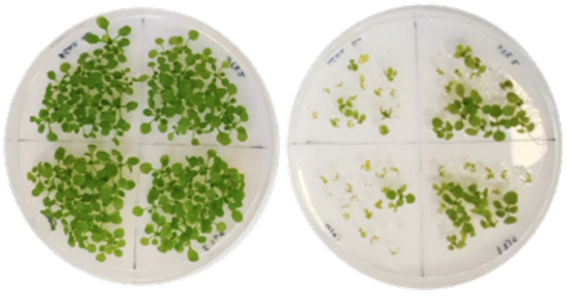
Flipping the switch; regulating protein synthesis in response to stress
In plants, like in most eukaryotes, the p70 ribosomal S6 kinases (S6Ks) pathway coordinates cell growth, cell proliferation, and stress response via modulating protein synthesis and ribosomal biogenesis. Studies in Arabidopsis thaliana (Arabidopsis) have shown that, similarly to in humans, the S6K family is composed of two members, called AtS6K1 and AtS6K2. Initial evidences suggest that AtS6K2 regulates responses to stresses and developmental cues. In fact, AtS6K2 is up regulated in response to cold and high-salinity stress and is co-expressed with genes involved in stress responsive regulation of plant growth. Furthermore, AtS6K2 plays an important role in chromosome stability and functions as a repressor of cell proliferation. In the context of future climate change predictions, we aim to understand how S6K2 modulates cellular responses to changes in the external environment in the model plant Arabidopsis. As such, our aim is to define how AtS6K2 allows plant cells to adapt to different environmental conditions. Plants are ideal to characterise S6K2 specific pathways, since our studies suggest that plant S6Ks represents the prototypical ribosomal kinase family.
Collaboration with Dr Ulrike Bechtold (Durham University)